How reliable is our genealogy evidence? We ask that question about evidence we mine from documents. When we use DNA, we should consider the reliability of our genetic evidence too.
What is “evidence” in genealogy? It’s the tentative answer to a research question; a hypothesis. Suppose our question is ‘Who were Margaret’s parents?’ We may find evidence in her baptismal record (where it’s likely very reliable) or we may find it on her death certificate when her son-in-law was the informant (where the answer may be less reliable). The less reliable source may be correct, but the informant may never have known his mother-in-law’s parents—he might be guessing or remembering wrong.
We have guidelines on evaluating traditional evidence. Is the source original or derivative? Is the information primary or secondary? Etc. But we don’t really have a consensus yet on guidelines for our DNA evidence.
Genetic evidence can also contribute to a hypothesis, but it uses DNA matches instead of documents. I’m still exploring how to evaluate the reliability of my DNA evidence. By reliable here I mean not just assessing the validity of two matching DNA segments, but a confidence level about how well that DNA match supports the tentative answer to the research question. Right now, I’m trialing a ten-point checklist for autosomal DNA (and X-DNA, which is included in most autosomal tests, but not Y-DNA or mtDNA.) Some of these steps may occur while I’m initially reviewing my matches, before I’ve even come to a hypothesis.
Two important notes: The sequence can vary! and Not all checklist points are always applicable!
I ask each question and then make note of my answers, even if they are “I don’t know” or “N/A”. This checklist gives me an opportunity to assess and record how effective or less-than-effective (or even conflicting!) any given DNA match might be in supporting my hypothesis.
Here are my ten points:
- Are our trees complete?
- Are our trees accurate?
- Does the amount of DNA shared by the two matches fall into the expected range for their hypothesized relationship?
- Are the individual matching segments large enough to be reliable?
- Do the matching segments fit as expected in my Visual Phasing chromosome map?
- Do the matches triangulate?
- Are the matches in a shared network?
- Can I walk the ancestors back?
- Is X-DNA a factor?
- Are admixture results a factor?
Let’s walk through an example. Because it’s March, and I always like to talk about Ireland in the month of St. Patrick’s Day, I’ll use an Irish research question!
INTRODUCTION TO THE FLYNN CASE
Research Question: Where in Ireland was James Flynn, son of Cormick Flynn and Alice Kiernan, born ca. 1836?
Background: We always need traditional evidence too. See Blaine Bettinger, “The DNA era of genealogy,” The Genetic Genealogist blog post, 17 Dec 2016 (https://thegeneticgenealogist.com/2016/12/17/the-dna-era-of-genealogy/). Only four men named Cormick Flynn headed a household in surviving Tithe Applotment (TA) books ca. 1830. These are a type of tax record; one of the few pre-Famine government sources extant in Ireland. I’ve compared the location of these four men to locations where a Kiernan family also appears in the TA records, and narrowed the search results to one county. (My report to myself contains all my research analysis and citations, but this blog post is limited to the DNA analysis.)
Tentative answer: The Cormick Flynn family lived in County Leitrim, Ireland, in the 1830s.
Figure 1 depicts the DNA matches I’m evaluating. This format is a McGuire Chart. See Lauren McGuire, “Guest Post: the McGuire Method – simplified visual DNA comparisons,” The Genetic Genealogist blog post, 18 Mar 2017 (https://thegeneticgenealogist.com/2017/03/19/guest-post-the-mcguire-method-simplified-visual-dna-comparisons/).
Figure 1.

On the left are DNA testers L1, L2, and L3. Each has ancestors born in County Leitrim, identified by green circles, and those lineages include the Flynn surname. To the right are DNA testers Ann and 3C (her third cousin) and 3c1r (their third cousin once removed), who each descend from different children of the immigrant James Flynn. The data on the colored lines on the bottom shows how much DNA each tester shares with the other testers. For space considerations, I’ve omitted the data that shows how much the three descendants of James Flynn share with each other, but the results were consistent with values for third cousin and third cousin once removed.
This example also ignores the possibility that the common ancestor could be related to the wife of James Flynn, rather than James himself; the evidence to dismiss that possibility is also omitted to save space.
THE CHECKLIST.
1. Are our trees complete? Flynn is not an uncommon name, especially in Ireland. Half of all my ancestors alive in the 1830s were living in Ireland, in perhaps eight different counties. How can I be confident that I’m not related to L1, L2, and/or L3 via some other ancestor, with the Flynn name just a coincidence?
James Flynn was my great-great-grandfather. Do I know all the lines of my pedigree chart to the next generation? (If I have gaps in my tree, our common ancestor could be someone whose identity I don’t yet know.) The good news is, I do know all 16 of my 2x-great-grandparents and 31 of my 32 3x-great-grandparents.
The bad news is, I can’t say the same for my matches. Some of them have limited online trees and/or haven’t replied.
There are ways to ameliorate the risk that tree completeness is less than ideal. For example, L3’s father was born in County Leitrim. If L3’s maternal grandparents were born in, say, Italy (and my tree has no Italian roots), and if L3 believes her ancestors lived in the same general areas for generations, then even if she doesn’t know half her tree in the target generation, it’s reasonable to assume we match on her County Leitrim side and not her unidentified-but-Italian side.
If I can take actions to improve the completeness of our trees, I will. Regardless, I make a note of our status on this checkpoint, so I can assess reliability when I look back at my evidence later, and move on.
2. Are our trees accurate? Cousin L2’s tree says her ancestor John Flynn was born in County Leitrim but died in New York. L2 found the County Leitrim information in online naturalization papers. But John Flynn is a very common name; do we know for sure that he was L2’s great-grandfather and not some other John Flynn? And what is the evidence that L2’s Flynn great-grandfather was in fact named John?
In this case, L2’s research was sourced and very solid. But with L1 and L3, I did research myself to validate and expand their trees. Then I make a note on this status—documenting the sources too—and move on.
3. Does the amount of DNA shared by the two matches fall into the expected range for their hypothesized relationship?
For Ancestry, 23andMe, MyHeritageDNA and gedmatch genesis, I use the totals they provide, which exclude very small matching segments. For FamilyTreeDNA, I count the total of matching segments that are at least 7 cM. Then I use the interactive Shared cM Tool developed by Jonny Perl, based on crowd-sourced data by Blaine Bettinger, available at DNApainter (https://dnapainter.com/tools/sharedcmv4) to determine potential relationships. That webpage also has a link called [More about this project] that leads to a 30+ page PDF with histograms and more detailed information, esp. about the lower probability results. In this example, I want to consider if the ancestors of L1, L2, and L3 could be first or second cousins to James Flynn. See Table 1 for a subset of that analysis.
Table 1.

I include this chart in my notes. Those hypothesized relationships fall in a valid range; two are better than average. It’s not conclusive of their degree of relationship, but it may help my hypothesis
- Note: If endogamy or pedigree collapse is involved, this can affect predictions. See Paul Woodbury, The Legacy Tree Genealogists’ Blog, “Dealing with Endogamy, part 1” (https://www.legacytree.com/blog/dealing-endogamy-part-exploring-amounts-shared-dna) and “Dealing with Endogamy, part 2” (https://www.legacytree.com/blog/dealing-endogamy-part-ii-test-multiple-relatives.)
4. Are the individual matching segments large enough to be reliable? Many people use different guidelines for this. In a study of my own family, I’ve found that matches over 15 cM appear genuine. See Ann Raymont, “When is a match a false positive?” DNAsleuth blog post, 8 Jul 2016 (https://dnasleuth.wordpress.com/2016/07/08/when-is-a-match-a-false-positive/.) For matches between 10-15 cM, I found 95% were valid. But 25% of matches between 7 and 10 cM appeared to be ‘false positives’. (My son had hundreds of matches in that range that matched neither of his parents. And most segments smaller than 7 cM failed to match either parent.)
For more information on small segments, see Blaine Bettinger, “A small segment round-up,” The Genetic Genealogist blog post, 29 Dec 2017 (https://thegeneticgenealogist.com/2017/12/29/a-small-segment-round-up/.)
Does this mean we can never use smaller segments? Like the example of the death certificate to identify the parents of the deceased, just because the evidence is less than ideally reliable doesn’t mean it’s wrong. But it does mean more work. Here are some ideas I’m exploring to assess reliability of segments between, say, 8 and 15 cM. (I still ignore any smaller segments.)
- I may want to see if the match appears in a “pile-up” region, which is “likely to be attributed to a shared ethnic history rather than recent common ancestry.” See ISOGG wiki, “Identical by Descent”, (https://isogg.org/wiki/Identical_by_descent). Ancestry’s algorithms weed these out, but other testing companies may present us with a cousin where our matching DNA is from hundreds of years ago. For a visual aid, see Debbie Kennett, “Small segments and pile-ups, a visualization,” Cruwys News blog post, Jan 2018 (https://cruwys.blogspot.com/2018/01/small-segments-and-pile-ups.html.) For a discussion of how it may affect match reliability, see Jim Bartlett, “Pile-ups,” Segmentology blog post, 7 Oct 2015 (https://segmentology.org/2015/10/07/pile-ups/).
- Phasing: if you have DNA from a parent as well as yourself, you can use phasing to improve the accuracy of your smaller matches. This is a complicated topic; I’ll refer you to Blaine Bettinger, “The effect of phasing on reducing false distant matches,” The Genetic Genealogist blog post, 26 Jul 2017 (https://thegeneticgenealogist.com/2017/07/26/the-effect-of-phasing-on-reducing-false-distant-matches-or-phasing-a-parent-using-gedmatch/longphase). AncestryDNA and MyHeritageDNA reportedly phase the data with other means before they report matches. See ISOGG wiki, “phasing” (https://isogg.org/wiki/Phasing).
- I don’t have a parent’s DNA, but I do consider other close kin who may also match my new cousin. For example, what if I match a new cousin “L4” for an 8.5 cM segment on FamilyTreeDNA? If my brother also matched L4 on that segment, would that make my match more trustworthy? What if my brother tested at 23andMe, and we’re comparing on gedmatch genesis? Does that reduce the risk that my FTDNA match to L4 was a genotyping error? What if my brother’s match is bigger, say 16 cM? Or 8.5 cM but he matches L4 on another segment as well? Do those situations make my 8.5 cM match more reliable?
To be honest, I don’t know the answer to this question, but I record all these observations in my project report to myself. This match to L4 may be more reliable than if none of my close family match him.
Fortunately, my matches to L1, L2, and L3 are 18-40 cM. But the matches between the County Leitrim kin and 3C and 3c1r may be less reliable. I don’t administer those kits, so for now I make a note of this and move on.
5. Do the matching segments fit as expected in my Visual Phasing chromosome map?
I am fortunate to have successfully completed Visual Phasing on my 23 chromosomes, using the DNA of several siblings to map which segments I got from each of my four grandparents. I wrote about Visual Phasing at Ann Raymont, “10 Tips to Trial a Tool,” DNAsleuth blog post, 1 Jan 2019 (https://dnasleuth.wordpress.com/2019/01/01).
Visual phasing tells me if the chromosome segment I am evaluating came to me from the grandparent I expect. In this case study-in-progress, my matches passed this checkpoint. For example, Figure 2 shows how the DNA I share with County Leitrim cousin L2, on chromosomes 11 and 15 (expanded below), did come to me from my paternal grandfather, as expected.
Figure 2. Visual Phasing, chromosomes 11-15, with L2 added

For the lineage that illustrates that I descend from James Flynn through my paternal grandfather’s line, see Figure 1 in the Introduction. For confirmation that this DNA segment was on my paternal copy of chromosome 15, see Table 2 below.)
6. Chromosome triangulation means that I have at least three testers descended from a common ancestor who all share an identical DNA segment (on the same location on the same chromosome), which was passed down to each of those testers from that common ancestor (ideally from different children.) I have blogged about the value of triangulation at Ann Raymont, “Triangulation with Gedmatch,” DNAsleuth blog post, 7 Mar 2017 (https://dnasleuth.wordpress.com/2017/03/07/triangulation-with-gedmatch/.) Triangulation requires a chromosome browser (something AncestryDNA doesn’t offer), so it’s not always an option. And even when the tools are available, groups of cousins could be descended from the same common ancestor without triangulating. Many genealogy experts using DNA (but not all) believe triangulation makes your case stronger. If I have evidence of triangulation, I include it in my notes.
In this case, I do have a triangulated segment on chromosome 15, shown in Table 2.
Table 2.
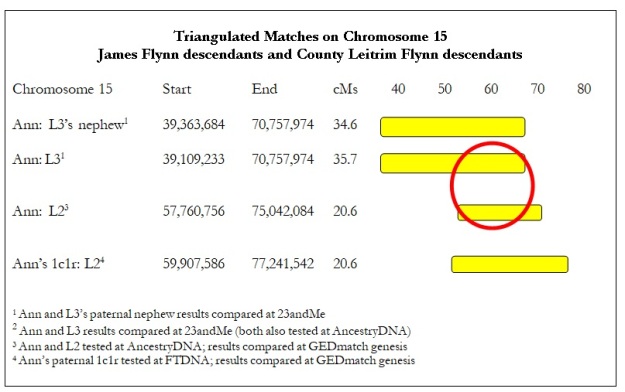
Ann (me), L2, and L3 triangulate. It’s less than 20 cM but likely more than 15 cM. Table 2 also includes a paternal nephew of L3 and a paternal first cousin once removed (1c1r) for Ann. Such close relatives don’t form independent ‘legs’ in a triangulation group, but they do strengthen the evidence that the match is on Ann’s paternal grandfather’s line and L3’s paternal line. (See Figure 1, in the introduction.) This triangulation group confirms that Ann, L2, and L3 share a single common ancestor, but it doesn’t prove how far back. In fact, a segment of only 20 cM could be 10 generations back. (See the section discussing the 2015 Speed and Balding paper at ISOGG wiki, “Identical by Descent,” (https://isogg.org/wiki/Identical_by_descent).
I also have a triangulated group for L3, 3C, and 3c1r on chromosome 3, but those segments are 11 cM or less. I include that group in my notes with a caveat about the segment size, and move on.
7. Shared networks can help when there is no chromosome data. Here, you have groups of genetic cousins, descended from multiple children from the same common ancestor. (In this case study-in-progress, I’m not actually identifying a specific shared ancestor; my research question is for location, not name. But the principle is the same.) I can include L1, who tested only at Ancestry, by comparing her to others who tested at Ancestry. See Figure 1. Among the Leitrim kin, I know she shares more than 20 cM with L3 and with me because she shows up on Ancestry’s Shared Match List for us. I have a precise amount (42 cM) shared between L1 and L2, because I asked L2. (L1 never replied.)
Why does it help to have a network or a triangulation group, instead of just one match? If you have just one match pair descended from the Most Recent Common Ancestor (MRCA), you may have evidence supporting your hypothesized MRCA. But if you have seven match pairs, or 17 match pairs (!), descended from several different children of the same MRCA and who mostly match each other too, your evidence is progressively more reliable; you can have more confidence in your tentative answer to the research question.
How many match pairs do we need to have strong, reliable evidence? Like much else in genealogy, it depends. The more distant the MRCA, the more pairs you want. (And beyond 5-7 generations, it’s very difficult to be confident the MRCA isn’t someone in an unidentified or unproven slot in folks’ pedigree charts instead of who you want it to be.) Also, the weaker the documentary evidence, the stronger the DNA evidence may need to be.
8. Can I walk the ancestor back? I first heard this phrase from Jim Bartlett, author of the Segment-ology blog (https://segmentology.org). I don’t focus only on specific chromosome segments, but I do make note if I can ‘walk the ancestor back’ generation by generation. For example, in this Flynn case, I have at least one first cousin, second cousin, and third cousin who descend from James Flynn and match one or more of the Leitrim kin. I also have at least one fourth cousin, descended from James Flynn’s brother Hugh, who matches me and Leitrim cousin L3 for more than 20 cM at Ancestry. (Fourth cousins are omitted from Figure 1 for space reasons.) This gives me more confidence that these matches reliably lead to a common ancestor in Ireland.
9. Is the X chromosome a factor? The X chromosome is not quite the same as chromosomes 1 through 22. Testing companies look at significantly fewer markers on the X chromosome than other chromosomes of the same size, so there is a greater risk of false positives, especially if one of the matching pair is a female. On the other hand, the X chromosome inherited by a male doesn’t go through recombination, so it as the benefits of a phased chromosome. At the Genetic Genealogy Tips and Techniques Facebook [closed] group in August 2018, Blaine Bettinger suggested that an X match between two men may be worth considering at 7 cM or higher. A match that involves a female will experience a higher number of false positives until you get to 15 cM or better still, 20 cM. So, I don’t spend time on X matches lower than these thresholds.
This Flynn case study does not include any X matches. If it did, I would want to be sure the lineage path to the hypothesized common ancestor follows the path of X chromosome inheritance. See Blaine Bettinger, “Unlocking the Genealogical Secrets of the X chromosome,” The Genetic Genealogist blog post, 21 Dec 2008 (https://thegeneticgenealogist.com/2008/12/21/unlocking-the-genealogical-secrets-of-the-x-chromosome/).
10. Admixture results (sometimes called ‘ethnicity’, or biogeographic origins) may only rarely come into play.
As Blaine Bettinger has said, “while ethnicity estimates have MANY limitations of which we must be aware, it is important to remember that they cannot be ignored. Despite their limitations, ethnicity estimates are still genealogical evidence and ignoring *any* evidence (especially evidence poured into your lap!) equates to poor research skills…. As long as we understand their limitations, ethnicity estimates are not solely entertainment.” (Source: Genetic Genealogy Tips and Techniques Facebook post 26 Feb 2019.)
Suppose Andy, an adoptee, is 50% Ashkenazi Jewish and his birth mother is not at all Jewish. If a DNA match is also Ashkenazi Jewish, this can make the match more valuable as supporting evidence for the identity of his birth father.
In this case study-in-progress, I have no data here yet to improve the reliability of my DNA evidence. I checked to see if County Leitrim is included in Ancestry, “List of DNA Regions” (https://support.ancestry.com/s/article/DNA-Regions). Leitrim is a subset of their Central Ireland region, but it’s not one they’ve associated yet with my DNA. I’ll make a note of that and perhaps continue to monitor it. (It’s not proof that I had no ancestors from County Leitrim; but it’s certainly not evidence that I did.)
CONCLUSION.
For every brick wall I work on, I write a report to myself, with timelines and genealogical summaries, maps and document transcriptions and abstracts, and analysis and correlation, resolving conflicts, and suggesting next steps. These reports have a DNA section too, in which I include ideas for additional targeted testing, a correspondence log, DNA test results, and an assessment of those results based on some of the factors I’ve outlined above. I also add specifics, such as GEDmatch kit numbers and proof supporting the testers’ lineages, which I’ve omitted here for privacy reasons. It can be a good idea to include screen captures of results too (match info and their trees), in case a tester later deletes his info or goes private.
Looking at this Flynn project, I can see that I need more evidence. I’ll continue to search for documentary clues. I am trying to recruit Y-DNA testing candidates, and I monitor my DNA matches. When new matches pop up, it’s helpful to have this data all assembled in a report to myself so I can pick up where I left off.
This blog post doesn’t guide you through preparing a proof argument that incorporates DNA. The National Genealogical Society Quarterly has many issues that provide examples, and Board for the Certification of Genealogists is revising the booklet Genealogy Standards this year to include updates that address DNA.
Rather, this is a snapshot of the questions I ask about my DNA matches today as I consider how to assess their value when trying to solve brick walls. It’s still an evolving process.
Happy St. Patrick’s Day!
Ann Raymont, CG® © 1 March 2019
All websites in the article were accessed 24 February 2019. My thanks to Shannon Green, CG®, Jan Joyce, CG®, and Sandra Maciejewski Porter for their feedback on an earlier draft.
To link to or cite this article: Ann Raymont, “How reliable is our DNA evidence?” DNAsleuth blog post, 1 Mar 2019 (DNAsleuth.wordpress/com/2019/03/01.)

This is very timely as I believe I’ve finally figured out who my paternal grandmother’s biological father is but I’ve been struggling in considering how confident I am or should be based on the DNA evidence. Her closest DNA match is a first cousin once removed and based on what I know of the family (limited remaining living family members/descendants) this may be as close as I can get. Now I feel that I’m at at least considering the right factors so thank you. Do you have any suggestions as to how many triangulated segments are enough? I’ve found at least 10 segments (15 cM or more) on different chromosomes that I can triangulate to her grandparents. The fact that they only have one son was my saving grace in this. My “gut feel” is that I’ve solved the mystery but I’d feel horrible if I was wrong and put this in my public tree!
LikeLike
That’s a great—and complicated—question! You may very well have solved your mystery, but I’ll throw a few comments out for consideration.
First, to address your specific question about how many match pairs you need—I haven’t seen any consensus on an answer to that. I know experts are discussing it, but there are many other variables intertwined, so I don’t know if or when we’ll see guidelines on that.
If I am reading you right, your paternal grandmother matches someone you’ve hypothesized is her 1c1r, based on other documentary evidence (ages, locations, etc.); is that right? Is there any other relationship that match could be, based solely on DNA? For example, a match of 150 cM could be a 1c1r, 2c, 2c1r, 3c, or 3c1r. A match of 540 cM is really only likely to be a 1c1r.
I just watched Blaine Bettinger’s presentation “Essential Considerations for DNA evidence” at rootstech.org (which I believe may be made available on that website in the near future), and he talked about making genealogy more ‘science-y’ and the importance of trying to disprove our hypotheses, as scientists do.
So you may want to see if there is *any other* ancestor of your grandmother’s matches that could be the shared ancestor, starting with other relationships suggested by the Shared cM tool. You can then try the ‘What Are the Odds’ tool at DNApainter.com to see the probabilities. (Type WATO in the search box of my blog if you want to read more about that tool.)
Finally, you mention caution in adding your conclusion to your public tree. I’m glad you brought that up. If there are living people who would be upset see to your discovery, that’s something to consider, especially if there is *any* doubt. I use an icon/image that says “Not Proven” when I want to include someone in a public tree, but it’s still a hypothesis. But I confess, I haven’t had to make that decision about someone in a recent generation.
Congratulations, and/or good luck! ~ Ann
LikeLike
Thank you for your reply Ann. I watched Blaine Bettinger’s presentation today too! Two great sources of information and inspiration in one day! Woo hoo!
To expand on my previous comments in case that’s helpful to anyone, my grandmother was born in 1918 so what helped me significantly was the DNAPainter shared cM tool you suggested. It took me a while to grasp that I was looking at potential common ancestors back about 1800 when really her DNA matches were once, twice or more times removed and several connected up with the couple that appear to be her paternal grandparents who were born closer to 1870 or two generations later than I initially expected.
At this point I have two matches of 1st cousins 1x removed (if the father is actually the father) and three 1st cousins 2x removed. I’ve now been able to confirm that the biological father has no other known living descendants. My grandmother has two living first cousins but one has significant overlap on her other side with my grandmother’s maternal side so determining the source of the segments would be extremely difficult. The other cousin has not committed to do a DNA test and if she doesn’t agree then my current closest matches will be the ones I will need to base a conclusion on. There are other more distant matches including a 1st cousin 3x times removed and one 5x removed and all share this same couple that I believe is my grandmother’s father. There are other matches that connect up to her great grandparents and further back too.
Fortunately both sides of my grandmother’s tree are filled in completely back to at least the early 1800’s (assuming my hypothesis is correct) so these DNA matches are sharing common ancestors within the realm of fully completed branches (which Blaine discussed extensively this morning) and also with good paper trails. I’ve also been fortunate in that I’ve been able to triangulate an X-DNA segment from my grandmother’s paternal side which came from her biological father’s mother’s line. I just checked and I’ve found triangulated segments (a few down towards 10 cM in size but most 15 cM or more) on 15 chromosomes + X which is probably as much as I’ve found on other testers where I have firm paper trails, so I sense that I shouldn’t be as hesitant as I am. I’m sure if I went back further and started looking at smaller segments I’d be able to fill in even more.
So I’m keeping my fingers crossed that the remaining first cousin will do a DNA test but I’m putting my ducks in a row as far as my notes anyway as I may have all the info I’ll get. I’ll also act on the advice to continue to prove ALL lines based on Blaine Bettinger’s assertion that sooner or later there’s probably an MPE. I received an email from a “cousin of a cousin” yesterday and I’ll be referring her to your blog to ensure that she’s covering all of her bases too as she’s in a similar situation. Thank you again for writing about this and then taking the time to look at my message.
LikeLike